Dr. Shen-Ying Zhang, Assistant Professor of Clinical Investigation at Rockefeller University is Co-PI with Dr. Jean-Laurent Casanova, Head of the St. Giles Laboratory of Human Genetics of Infectious Diseases, on two NIH (R01) research project grants. R01 grants are prestigious awards that provide support for health-related research and development aligned withthe mission of the NIH. Dr. Zhang is a graduate of the Clinical Scholars Program and a member of the Rockefeller Early Phase Physician Scientists (REPPS) group.
The first R01 is titled Cellular Dissection of Herpes Simplex Encephalitis with iPS Cells. It focuses on analyzing Herpes simplex virus 1 (HSV-1) encephalitis (HSE), the most common form of sporadic viral encephalitis in Western countries. The St. Giles Laboratory of Human Genetics of Infectious Diseases discovered that childhood HSE may result from inborn errors of Toll-like receptor 3 (TLR3)-dependent, interferon (IFN)-α/β-mediated immunity against primary infection by HSV-1. The lab has identified mutations in the TLR3-pathway molecules UNC-93B, TRIF, TRAF3 and TBK1 that are important in a cell’s ability to sense the presence of viral double-stranded RNA (ds-RNA) molecules. Children with mutations in STAT1 or NEMO have also been identified and they have broader impairment in immunity and are predisposed to both HSE and other infections.
Dr. Zhang’s research has shown that forebrain neurons and pro-oligodendrocytes derived from TLR3- and UNC93B-deficient induced pluripotent stem cells (iPSCs) have markedly abnormal cellular responses to poly(I:C), a TLR3 mimic of dsRNA, and are highly susceptible to HSV-1 infection, unlike other central nervous system (CNS)-resident cells tested. These data suggest that childhood HSE results from inborn errors of non-hematopoietic, CNS-specific, “intrinsic” immunity, affecting neurons and oligodendrocytes in particular. This novel finding changes the paradigm of immune compromise form impaired immune blood cell function to tissue-specific defects in combating viral infection.
The goal of her current research is to dissect in greater depth, and from three complementary angles, the neuron-intrinsic pathogenesis of HSE. First, a faster protocol will be devised to differentiate and test iPSC-derived forebrain neurons from healthy controls and four groups of patients, including those with mutations in (i) HSE-predisposing TLR3-pathway genes (TRIF, TBK1, TRAF3, NEMO), (ii) HSE-predisposing IFN-α/β- and -λ-pathway genes (STAT1 and newly discovered IFIT2), (iii) other IFN-pathway genes not related to HSE (IL10RB, TYK2, IRF7), and (iv) novel HSE-causing genes that cause immune defects by unknown mechanisms (DBR1, SNORA31). Second, a novel protocol will be devised to differentiate iPSCs into trigeminal neurons in which HSV-1 establishes latency in children without HSE, and compare control cells and cells with mutations in TLR3, STAT1, IL10RB, DBR1, and SNORA31 for their response to poly(I:C), IFNs, and HSV-1. Third, she will study the entry, retrograde axonal transport, gene delivery and replication of HSV-1 in forebrain and trigeminal neurons from patients with defects in TLR3 function. To demonstrate the disease-causing role of any genetic defect, isogenic iPSC lines will be tested in which the mutation has been corrected or introduced by CRISPR-Cas9 gene editing. Exciting preliminary data have already been obtained, including: (i) novel genetic etiologies of childhood HSE (IFIT2, DBR1, SNORA31), (ii) novel Sendai virus (SeV)-based generation of iPSCs, (iii) novel protocols to differentiate HSV-1-permissive forebrain and trigeminal neurons, and (iv) novel imaging of HSV-1 infection in neurons.
While studies of HSE have traditionally been limited to animal models, the pursuit of this human iPSC-based study will enable Dr. Zhang and her colleagues to dissect in-depth the molecular and cellular basis of HSE in children with inborn errors of CNS-intrinsic immunity to HSV-1. This path-breaking collaborative study has far-reaching medical and biological implications.
The second R01 grant is titled Mendelian Genetic Predisposition to Herpes Simplex Encephalitis in Childhood. It focuses on childhood herpes simplex encephalitis (HSE), which is a life-threatening complication of primary infection by herpes simplex virus 1 (HSV-1), a common virus that is typically innocuous. HSE is the most common sporadic viral encephalitis in Western countries and even with treatment with the antiviral drug acyclovir, survivors often suffer from severe neurological sequelae. The pathogenesis of HSE remained unclear until the research conducted by St. Giles Laboratory of Human Genetics of Infectious Diseases showed that the disease results, in some children, from single-gene mutations impairing TLR3- and IFN-α/β-mediated immunity to HSV-1 in the central nervous system (CNS).
Following a candidate gene approach, 13 patients were reported with rare mutations in one of the TLR3 pathway gene (TLR3, UNC93B1, TRIF, TRAF3, TBK1). In parallel, a hypothesis-generating search was initiated for novel gene defects that may predispose a patient to HSE by studying well characterized HSE kindreds by genome-wide (GW) linkage (GWL). Using this method, DBR1 was discovered as a novel HSE-causing gene in two relatives. However, no genetic etiology has yet been identified for 235 of the 250 HSE patients under study. The research team hypothesizes that HSE in some of these children is a consequence of a collection of CNS-intrinsic inborn errors of immunity to HSV-1, possibly but not necessarily related to the TLR3-IFN-α/β circuit.
The goal of this research is to extend the GW approach by taking advantage of whole-exome sequencing (WES) to both the 28 patients consanguineous and 207 non-consanguineous patients (trio design). The WES data will be analized in two ways: 1) hypothesis-based, searching for mutations in TLR3-IFN-α/β pathway genes; 2) hypothesis-generating, searching for mutations in other genes.
The research will benefit from GWL and human gene connectome analysis. Whole-genome sequencing (WGS) will be performed in patients for whom WES fails to reveal candidate mutation. Patients’ fibroblasts will be used to investigate the impact of the new candidate genetic etiologies on anti-HSV-1 immunity.
This grant is strengthened by the investigators developing an international cohort of 230 children with HSE, their finding rare mutations in 18 key genes of the TLR3-IFN pathway in up to 52 patients and their identifying by unbiased GW analysis rare heterozygous mutations in the IFN-inducible gene IFIT2 in 4 patient and in a small non-coding RNA gene SNORA31 in 6 other patients. The research will decipher the pathogenesis of a devastating pediatric illness, paving the way for new therapeutic approaches. The genetic analysis of HSE will also provide proof-of-principle that sporadic, life-threatening infectious diseases in otherwise healthy children may result from single-gene inborn errors of immunity.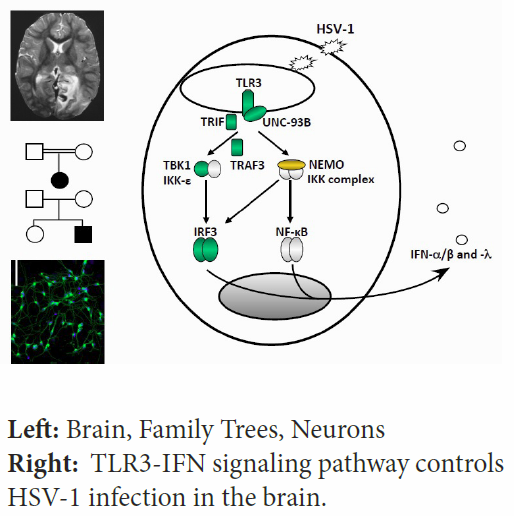
February 26, 2016
Clinical Scholar Graduate Dr. Shen-Ying Zhang is Studying the Genetics of Immune Deficiences with the Support of Two NIH RO1 Grants
By Editorial Staff